Announcing a massive innovation in protein Higher Order Structural analysis:
The New AutoFox® System
from GenNext
Hands-free, high-throughput, and reproducible protein footprinting


For the first time, researchers can harness the power of fully-automated, chip-based Hydroxyl Radical Protein Footprinting with the new AutoFox System.
Progressing from First to Second Generation Technology
Big leaps in functionality, performance, and ease-of-use
Comprised of a user-friendly benchtop instrument coupled with intuitive data analysis software, our second-generation instrument, the AutoFox System, has grown into a more robust and hands-free tool.
Designed for high-throughput applications, the AutoFox System is a powerful tool for antibody-antigen epitope and paratope mapping, drug target engagement and allostery studies, protein interaction analysis, protein aggregation studies, PROTAC and molecular glue studies, and many more.
Complete automation without operator intervention to process up to 48 different samples at the push of a button.
No need for finicky fluidic and optical connections with the new optofluidic chip.
No need to premix samples with H2O2 with on-board mixing on the optofluidic chip.
Samples can be labeled in seconds instead of minutes with an increased illuminated flash volume from 0.1 μL to ~ 3 μL.
Flash lamp intensity automatically adjusts for changes in background scavenging with real-time radical dosimetry closed-loop control.
Advancements for High-Throughput & Reproducible HRPF


AutoFox Optofluidic Chip
Plug-and-Play HRPF
At the heart of the AutoFox system is our proprietary optofluidic chip that eliminates the need for fused silica capillaries typically employed in HRPF studies. This compact, reusable microfluidic device is comprised of the following:
- on-board, high efficiency, microfluidic mixer for combining protein samples with labeling reagents,
- serpentine photolysis cell that labels up to 30 µL in 10 seconds, and
- integrated dosimetry cell that provides real-time assessment of effective radical load, ensuring robust, reproducible, and actionable results even in the face of varying background scavenging.
The self-aligning mechanism of the AutoFox optofluidic chip makes changing chips fast and simple. The chip is easily installed and removed from the system via a compact manifold that ensures proper fit to optical and fluidic components. Gone are the frustrating days of installing capillaries with finicky optical and fluidic attachments.
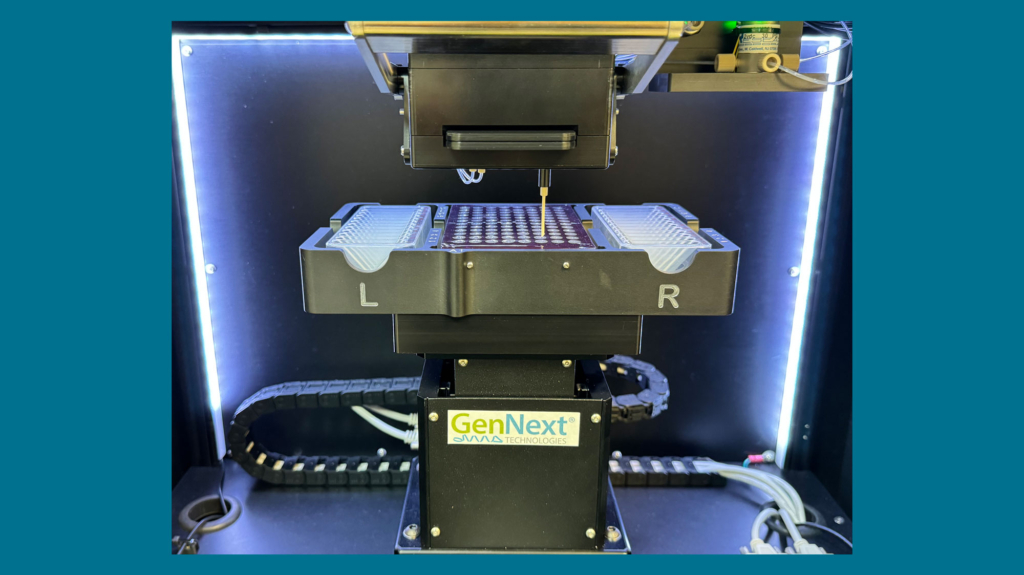

AutoFox Robotics
Automated labeling at the push of a button
The system’s automated, high-throughput capacity significantly reduces hands-on time and facilitates easy workflow integration by labeling each sample in 30 seconds, with the capacity to label up to 48 different samples.
The AutoFox sample deck is driven by a custom three-axis robot that moves samples, reagents, and quenching solutions directly to and from the optofluidic chip, automating full HRPF labeling and minimizing experimental error.
Samples and quench solutions are presented using a 96-well microtiter plate. Twenty-four additional 3 mL wells are provided for on-deck collection of waste and storage of chip wash solutions. The self-aligning chip is designed for precise registration of optical and fluidic trains and is easily installed or removed from the clamshell assembly.
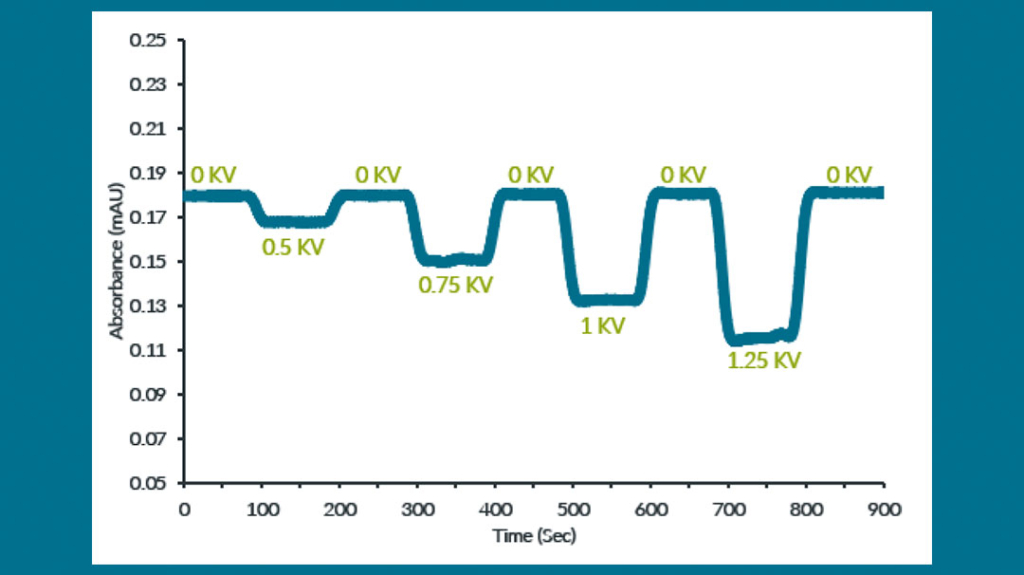

On-Chip, Real-Time Dosimetry
Closed-loop control of effective radical yield
Based upon proprietary radical dosimetry technology, the AutoFox System photometrically determines effective radical concentration in real-time and automatically adjusts flash fluence to achieve reproducible radical load even in the face of varying protein load, introduction of protein ligands, as well as variation in buffer composition and excipients. Real time dosimeter display enables easy inspection and validation of desired radical load.
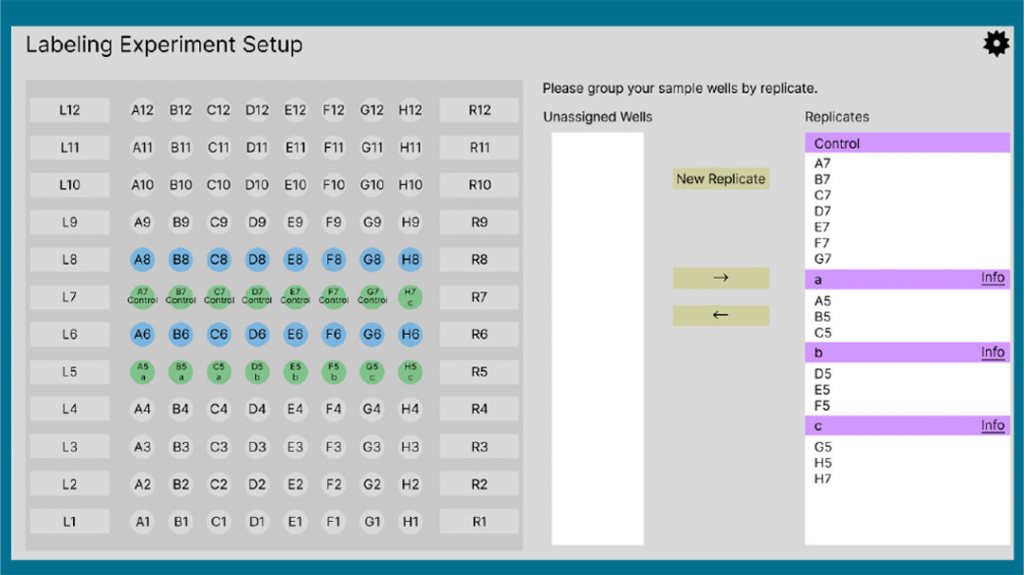

Intuitive Control & FoxWare® Software
Easy creation of method and sequence files, along with intuitive downstream data analysis
The AutoFox System is PC-controlled by an easy-to-use interface that runs on a laptop computer. Method and sequence file architecture makes for simple and reliable system programming and operation.
FoxWare Software is a powerful bioinformatics tool developed specifically to meet the demands of HRPF analysis. The software removes data processing bottlenecks and generates actionable results with a user-friendly interface and built-in analysis tools. Intuitive algorithms enable qualitative and quantitative comparative studies of the HOS footprint to address key requirements in biopharmaceutical and biosimilar research.



